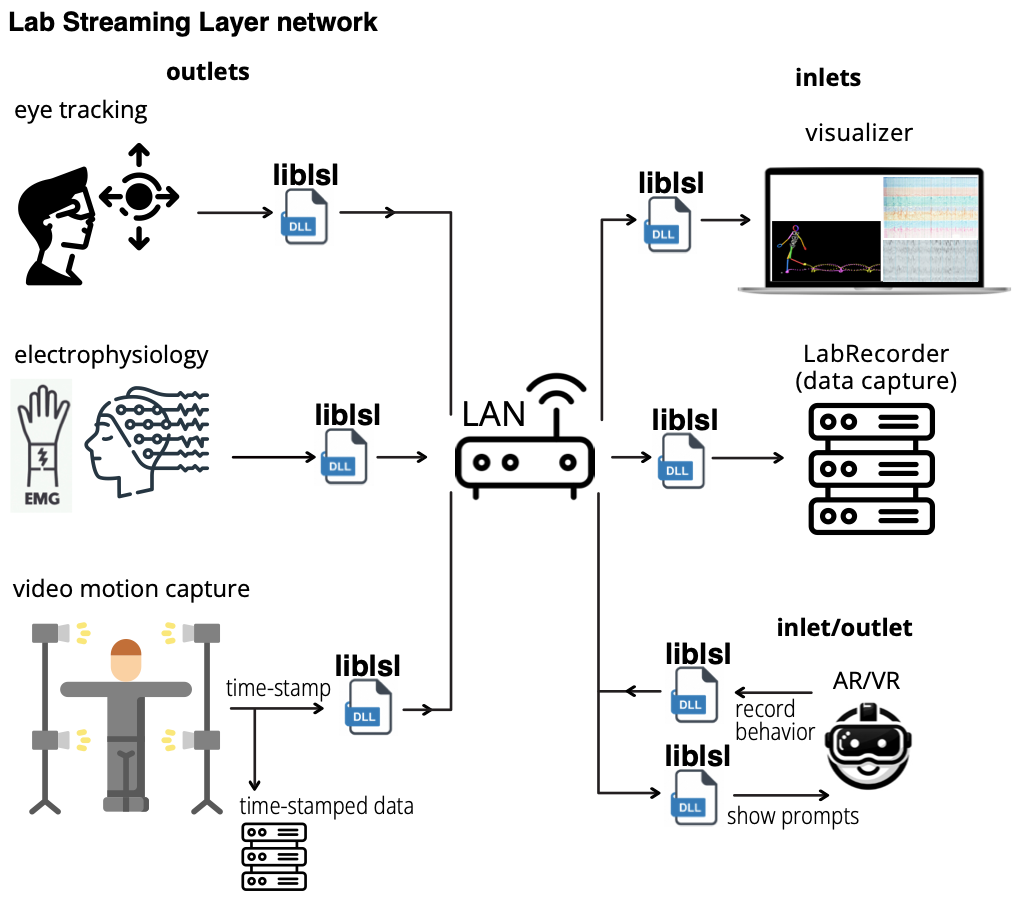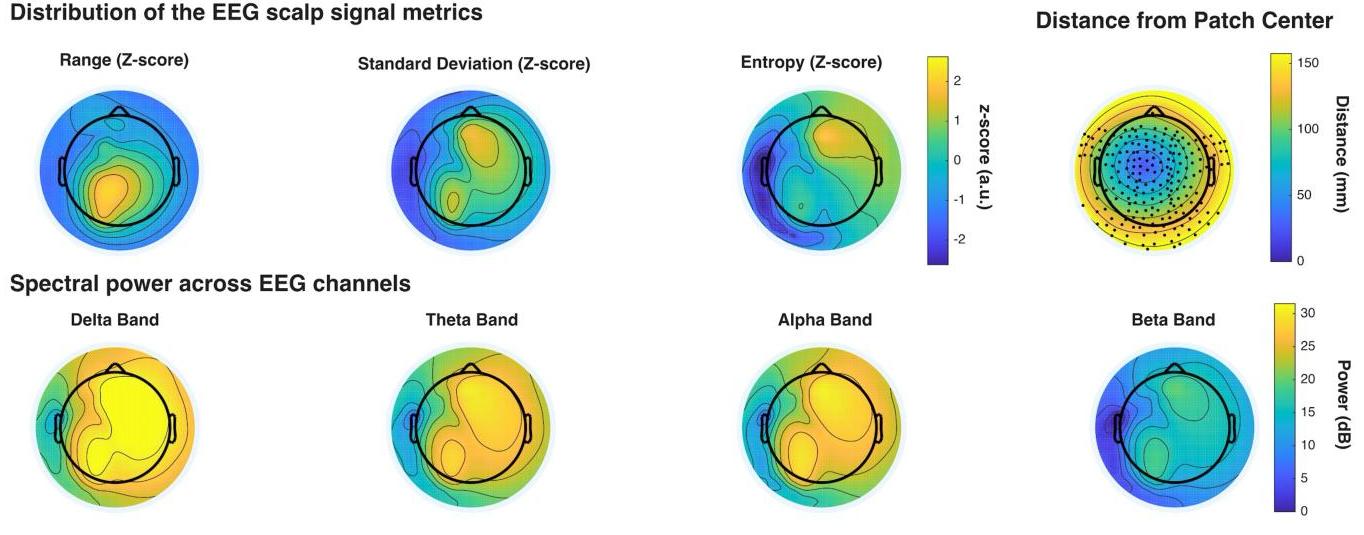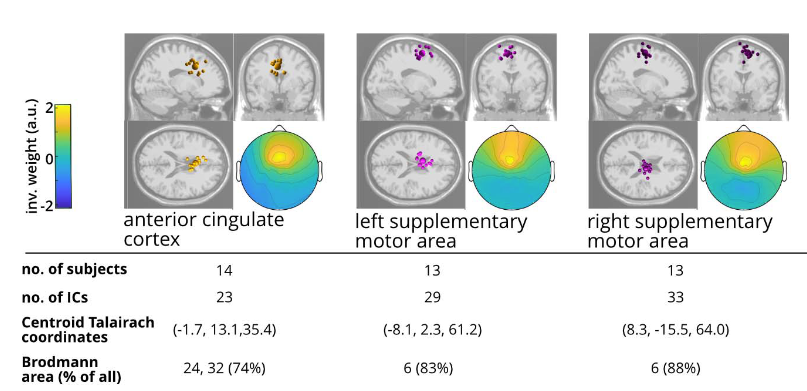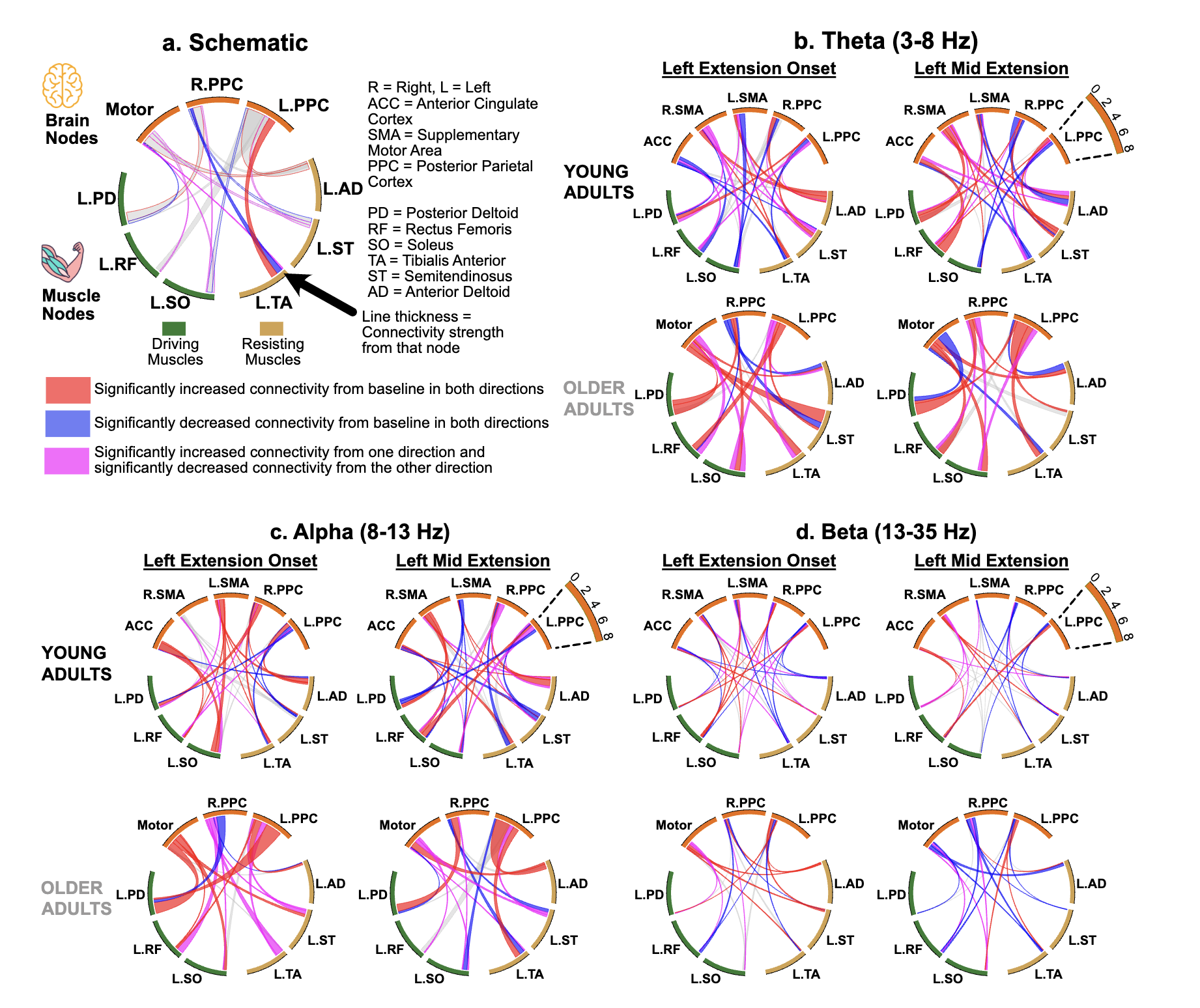
Age-related Reorganization of Corticomuscular Connectivity During Locomotor Perturbations
How does the brain communicate with muscles during unexpected perturbations, and how does this change with age? We investigated corticomuscular connectivity during perturbed recumbent stepping in young and older adults using high-density EEG and EMG. Young adults demonstrated selective connectivity between error-processing brain regions and specific muscles, with strong involvement of the anterior cingulate cortex. In contrast, older adults showed elevated baseline connectivity and relied on diffuse patterns dominated by motor and posterior parietal cortices, connecting to multiple muscles simultaneously regardless of their biomechanical role. This reveals a strategic reorganization: young adults use dynamic, error-driven control, while older adults employ a stability-focused approach that maintains comparable performance through constitutive hyperconnectivity. These distinct connectivity signatures establish perturbed recumbent stepping as a valuable tool for assessing age-related sensorimotor changes and developing targeted rehabilitation interventions.